Published on 27 August 2019
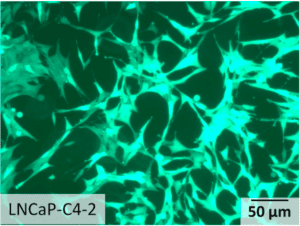 Figure 1 : Micrograph depicting the morphology of prostate cancer cell line named LNCaP-C4-2, expressing GFP.
Figure 1 : Micrograph depicting the morphology of prostate cancer cell line named LNCaP-C4-2, expressing GFP.
Prostate cancer is the second leading cause of cancer-related death for men. Circulating tumor cells (CTCs) are considered as a marker of early cancer diagnosis and disease severity. Their screening in blood is thus crucial to detect metastatic stage of cancer patients.
Nowadays, there is no reliable method available to capture and enumerate all CTCs because of their rare count in blood (1-100 cells/mL which is no more than 0,000002% of all blood cells) and because all current systems require an initial CTC isolation or require the use of antibodies. Another limitation is their reliance upon the expression of the surface markers EpCAMs (epithelial cell adhesion molecule). They are expressed on most cells within primary epithelial tumors, but their expression is often lost during tumor progression, so during metastatic stage.
Another approach to study tumor progression and cancer development uses the microfluidic organ-on-chip technology. Read here for more information.
To solve this challenge, the microfluidics community developed new CTC technologies that rely on biophysical attributes of CTCs. In particular, separation based on the larger size of CTCs compared to other cells in blood have been broadly investigated. Some microfluidic chips include constriction channels, micro filter arrays, or fan-in-fan-out micro cavity arrays to capture CTCs. However, these chips have limitations in throughput and capture efficiency.
At the light of the previous CTC systems, Xiang Ren et al. developed a new size-based CTC entrapment chip based on size separation offering better throughput and capture efficiency. The use of the OB1 Mk3 Flow Controller to capture prostate cancer cells (LNCaP-C4-2) with >95% efficiency, spiked in mouse blood at ~50 cells/mL.
APPLICATIONS
- High-throughput circulating tumor cells entrapment
- Accurate circulating tumor cells enumeration
- Early prostate cancer diagnosis
- New chip generation opportunities for other cell types (channel sizes, channels number, multiple chips connected…)
MATERIALS & METHODS
Materials
CTC high-throughput entrapment chip (CTC-HTECH)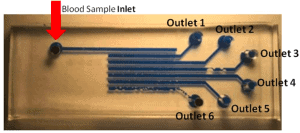
Set-up
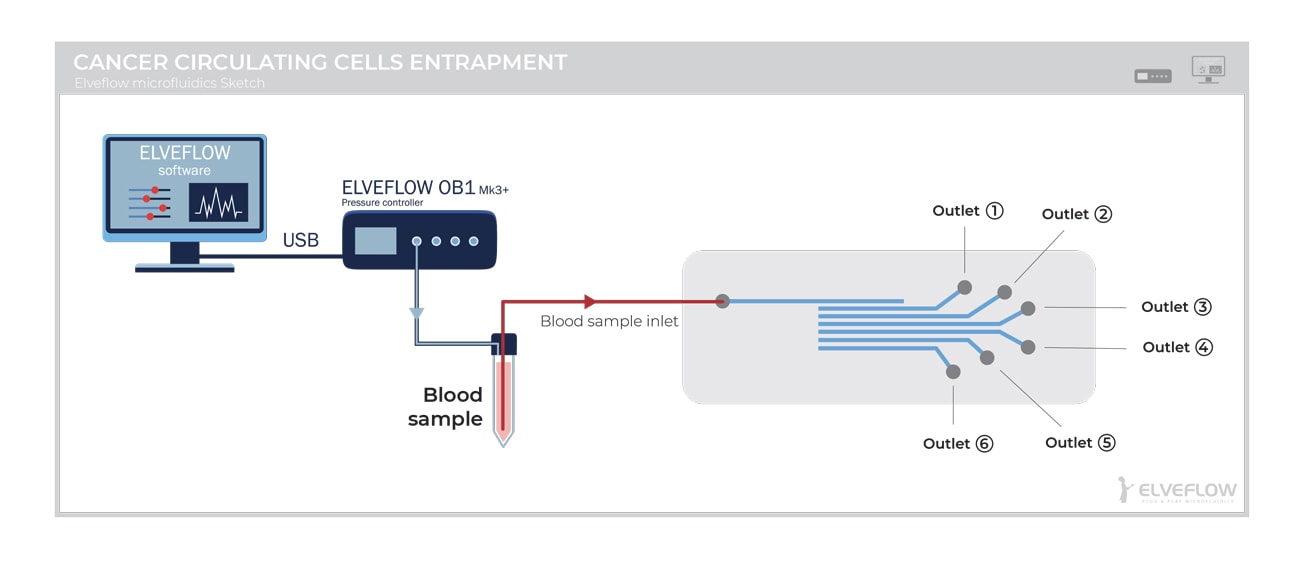
[starter_pack_rebound]
Methods
The CTC-HTECH chip is composed of one polydimethylsiloxane block (PDMS, polymer) obtained by soft-lithography and bonded to a glass slide by plasma treatment. The PDMS block contains a network of 6 straight parallel canals linked by a series of 40 constriction channels. Showed in the subsection “Materials”, constriction channels are also displayed on Figure 2, with an example of 2 parallel channels (red and blue).
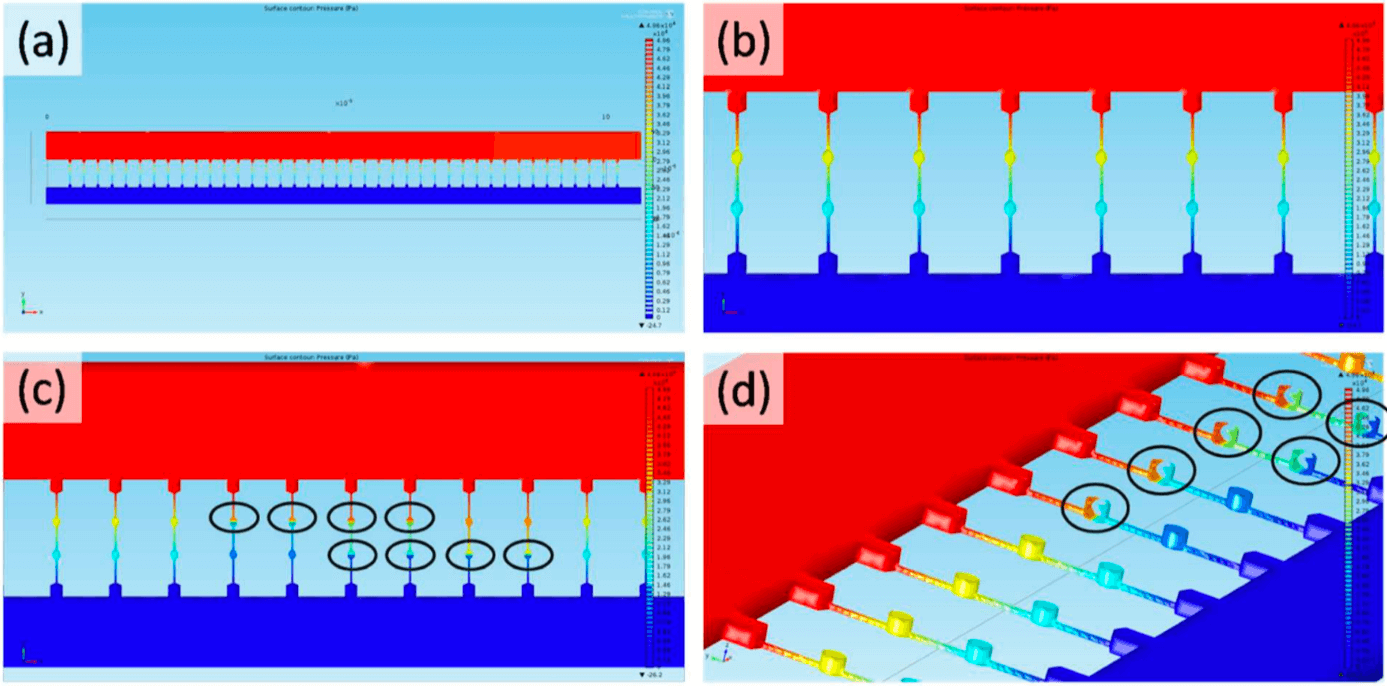 Figure 2 : Simulation of pressure distribution along the Circulating Tumor Cell-HTECH chip.(a) The pressure (Pa) distribution at row 1. The red color is associated to high-pressure while blue is for low-pressure.
Figure 2 : Simulation of pressure distribution along the Circulating Tumor Cell-HTECH chip.(a) The pressure (Pa) distribution at row 1. The red color is associated to high-pressure while blue is for low-pressure.
(b) Enlarged pressure distribution near microchannels
(c) Enlarged pressure distribution at row 1 with cells trapped in trapping chambers (highlighted in black circles)
(d) Enlarged pressure distribution near the microchannel with cells trapped in trapping chambers (channels with cells highlighted in black circles)
Each constriction channel is composed of 2 trapping chambers (Figure 2d). As cancer cells have a similar size to trapping chambers, they are deformed in the constriction channels and recover their shape within the cavities of the trapping chambers. The single Circulating Tumor Cells are captured in those trapping chambers, while the rest of the cells pass through the microchannels with the blood flow. After all of the blood sample has passed, chambers containing CTC are scanned (GFP fluorescence), allowing to determine the total number of CTCs. This unique feature is not available in other size-based CTC microfluidic chips (cf. Review CTCs).
A constant pressure of 500 mbar was applied to the blood sample reservoir using the OB1 Mk3 Pressure Controller. The resulting flow rate of the blood sample was ~ 2.4 mL/hour. By varying the supplied pressure, the OB1 pump maintains a constant liquid pressure in the system. This results in a variation of the flow rate as the cells become trapped in the microchannels but avoids excessive pressure that imposes additional mechanical stress and thus damages the cells (as syringe pumps do). Indeed, the syringe pump produces constant flow rate mode, guaranteeing the efficiency of most microfluidic devices with a limited number of microchannels. However, by increasing the numbers of microchannels in the CTC-HTECH chip, the pressure is redistributed, causing mechanical drag forces on the trapped cells, altering cytoskeleton strength, membrane stiffness, and biomechanical properties.
MICROFLUIDIC RESULTS
Figure 3a shows the CTC-HTECH device composed of the 6 rows ①-⑥ previously mentioned, where row ① is closest to the blood inlet port. The outlets 1-6 were defined as waste collection sites from rows ①-⑥, respectively. Only one outlet was opened during each trial. For example, when outlet 1 is opened, outlets 2 to 6 are closed, the Circulating Tumor Cells are trapped only in the trapping chambers of the row ①, while outlet 1 collects other blood cells (Figure 3a). When outlet 2 is opened, the CTCs are trapped in rows ① and ②, and outlet 2 collects other blood cells.
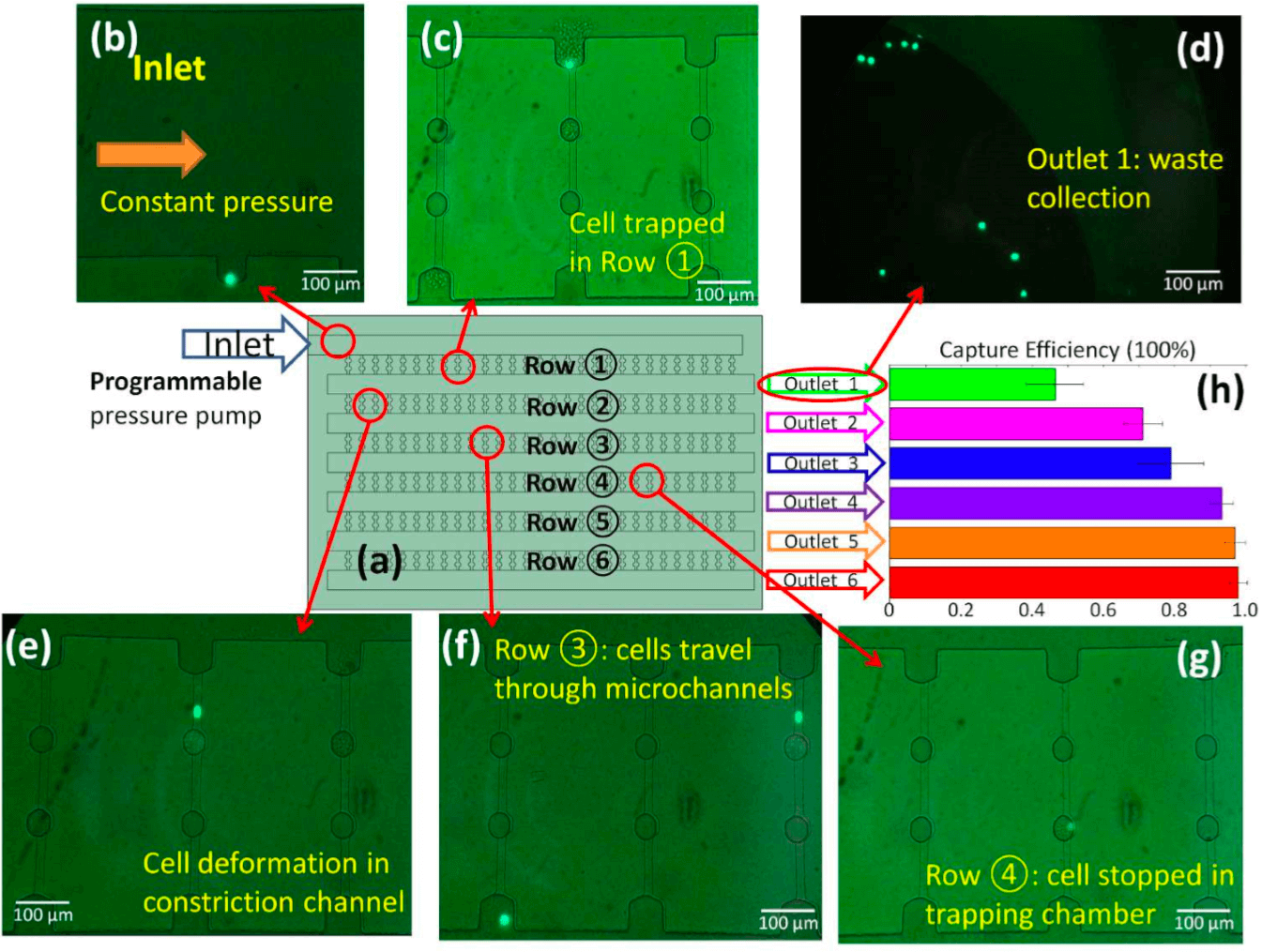 Figure 3 : (a) Illustration of the configuration of the device (not to scale) with the inlet connected to Elveflow’s OB1 Mk3 Pressure Controller ; each row and each outlet was assigned and labeled individually
Figure 3 : (a) Illustration of the configuration of the device (not to scale) with the inlet connected to Elveflow’s OB1 Mk3 Pressure Controller ; each row and each outlet was assigned and labeled individually
(b-g) The GFP+ LNCaP-C4-2 prostate cancer cells
(b) Image of inlet with a GFP+ cell starting to enter row 1
(c) GFP+ cell trapped in row 1 after the blood flow ceased
(d) Image of the waste collection at outlet 1
(e) GFP+ cell deforming and passing through row 2
(f) Two GFP+ cells in row 3 with one cell still passing, and one cell exiting this row
(g) GFP+ cell trapped in the trapping chamber of row 4
(h) Overall capture efficiency of each outlet. The data presented here is from 3 or 4 runs on CTC-HTECH device
The main conclusion of the previous figure is that when outlet 1 was opened and only row 1 was used for capture, the trapping efficiency was 46.3% (Figure 3h green bar) ; when outlet 6 was open and rows 1–6 were used for capture, the trapping efficiency was 97.9% (Figure 3h red bar). Five or six rows of micro constriction channels are needed in order to reach a capture ratio >95%.
The authors suggested that as the number of rows increases, the overall fluidic resistance also increases and, therefore, the trapping efficiency in each row varies. Thus, when only outlet 1 was used, the low fluid resistance led to higher flow rate which was too fast for all cancer cells to be captured. Therefore, using only a couple of rows will significantly decrease the number of trapped CTCs.
CONCLUSION
In this publication, Xiang Ren et al. presented a new size-based CTC entrapment chip increasing the capture efficiency . This low-cost system allows a trapping efficiency of 96%. This is significant as the number of Circulating Tumor Cells has been shown to be diagnostic or prognostic marker for tumor. The microchip run time was ~30 min for a 1.2 mL blood sample. Compared to magnetic particles with antibody methods, such as FDA approved CellSearch system, the CTC-HTECH reduced the lengthy sample pre-processing and long analysis time of ~4-6 hours.
The efficiency of this chip is guaranteed by the used of the OB1 Mk3 Pressure Controller. It can be used for CTC enrichment and entrapment for clinical diagnosis using liquid biopsies. As a customizable device, this chip can be extended following the same design for better capture efficiency, changing channel sizes, channels number, multiple chips connection…
References
- Xiang Ren et al. Entrapment of Prostate Cancer Circulating Tumor Cells with a Sequential Size-Based Microfluidic Chip. Anal. Chem. (2018)

Microfluidics knowledge
Do you want tips on how to best set up your microfluidic experiment? Do you need inspiration or a different angle to take on your specific problem? Well, we probably have an application note just for you, feel free to check them out!


 Job
Job Collaborations
Collaborations Customer
Customer Other
Other